-
 Our ResearchChromatin and systems biology
Our ResearchChromatin and systems biology
About our research
Epigenetic mechanisms and translational studies
ATP-dependent chromatin remodelers, such as SWI/SNF (BAF) and related complexes, play essential roles in regulating DNA accessibility. SWI/SNF activity is used by most cells to regulate key transcription factors. But how do these chromatin regulators choose where to make DNA accessible across the genome?
In our studies (Chambers et al., Cancer Research 2023), we found that SWI/SNF complexes are recruited to key enhancers by pioneer transcription factors. These pioneer factors bind their sites and recruit SWI/SNF chromatin remodeling activity to generate DNA accessibility needed by downstream transcription factors.
We are interested in understanding these mechanisms in cancer and in normal tissues. Surprisingly, inhibitors of SWI/SNF complexes are well-tolerated in vivo and pre-clinical studies show great promise for treating certain types of cancer. Basic mechanisms and translational studies focused around emerging SWI/SNF inhibitors represent major areas of interest to the lab.
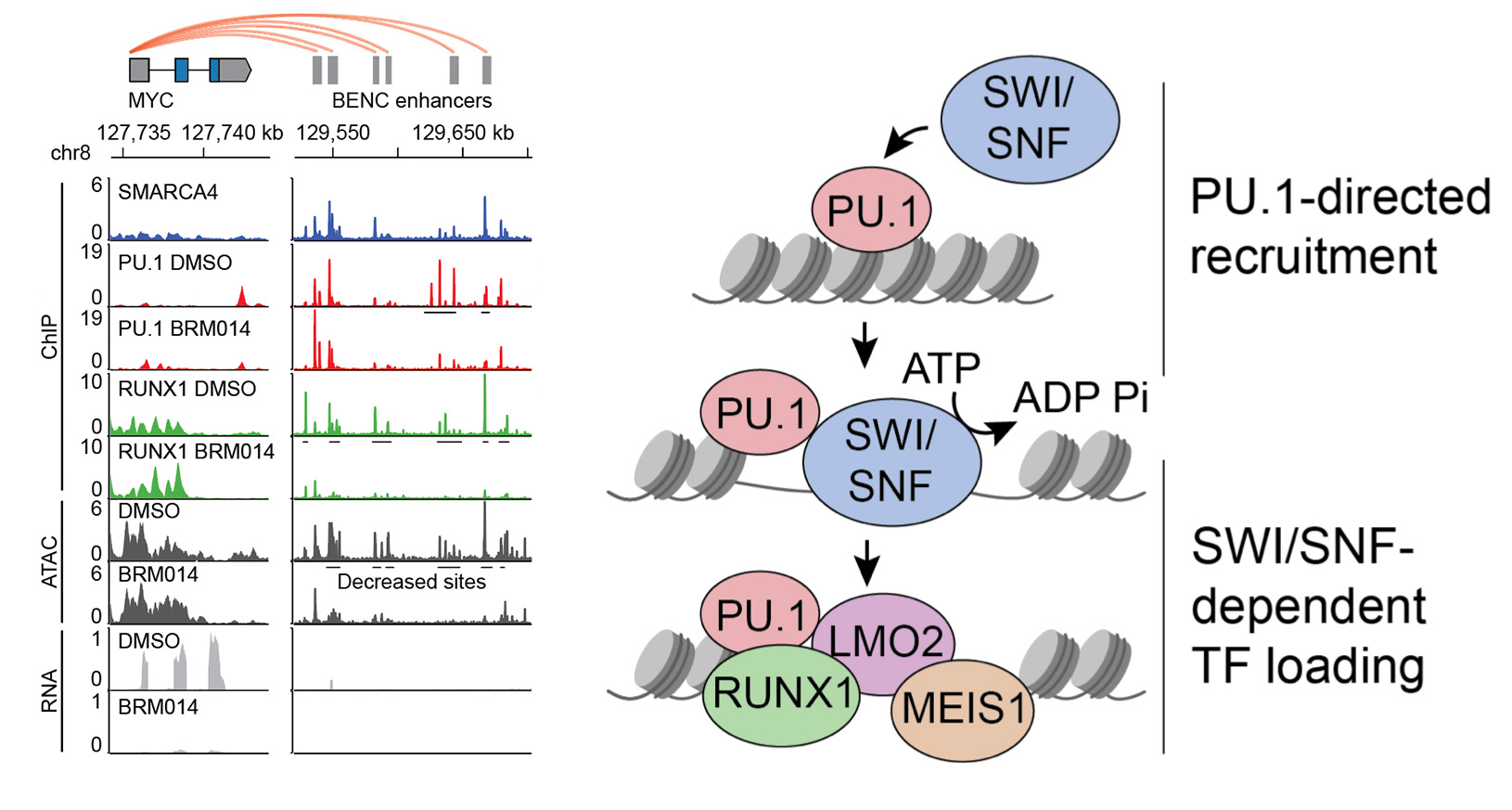
SWI/SNF activity is directed by the hematopoietic pioneer transcription factor PU.1. SWI/SNF inhibition deregulates PU.1 enhancers, which causes AML regression but also has important effects on normal PU.1-dependent cells.
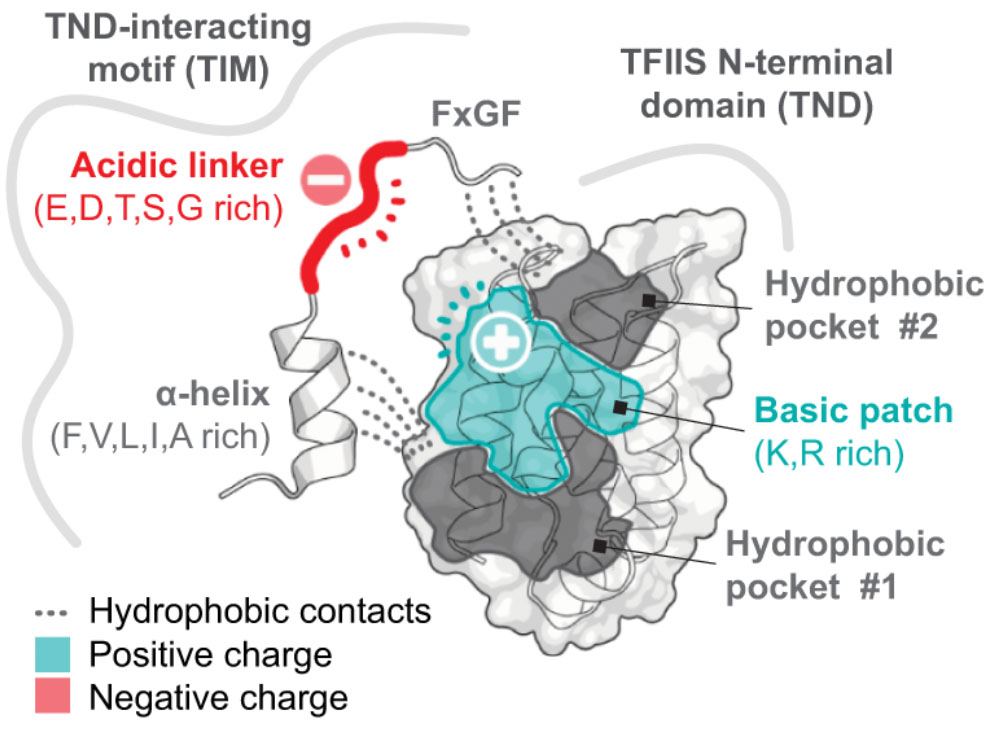
The roles of disordered protein in transcription regulation
Chromatin and transcription regulators are uniquely enriched in unstructured protein sequences referred to as intrinsically disordered regions (IDRs). IDRs in the gene expression machinery play many important roles, but what what mechanisms coordinate these disordered protein regions to influence transcription at specific genes?
In our studies (Cermakova et al., Science 2021), we identified a widespread domain that selectively reads IDRs. These domains or the motifs that they bind are found on most activating chromatin and transcription regulators, as well as major transcription factors. A growing number of these interaction modules are now known to play important roles in many settings. We are interested in understanding the spatial organization and the grammar by which IDRs coordinate to influence transcription.
Approaches we use in the lab
Our lab is a hybrid lab that integrates both wet- and dry-lab approaches. We are an interdisciplinary group with wide interests, and we use many approaches. Training in the generation and analysis of genome-wide data is at the lab's core. Trainees also develop skills in live-cell microscopy, single-cell analysis, chemical biology, translational biology, analysis of human specimens, and epigenetic methods. We also collaborate with clinicians throughout the Texas Medical Center for projects focused on specific malignancies. Upon project completion, trainees are ideally positioned for careers in industry or academia.
To read more about our research, see our publications.